-
Notifications
You must be signed in to change notification settings - Fork 154
Adding in many new surface families #368
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Conversation
|
It's generally better to have individual commits for each change rather than lumping everything into one big commit. It makes reviewing easier and the git log easier to understand. We would really appreciate it if you could re-commit your changes, e.g. with one family per commit. I have also made some changes to the new family drawings and re-ran the process_family_images.py script. Since I can't push to your branch, those are located on I assume @mazeau has been using these families for a while (?) and can confirm that they work as expected. It would be helpful to include more details in the PR message about what these families are, the data sources, and any results (even qualitative) you have from using them. For reference, #161 has a really great write-up (though that level of detail is not necessarily required). Finally, (perhaps @rwest could also comment) we haven't yet settled on how we want to approach reviewing of catalysis related PRs. For RMG-database, it seems that the best we can reliably do is check syntax and formatting and confirm that the database can be loaded. Ideally, the primary reviewer would be someone else who works on catalysis, but I don't know if there is anyone suitable currently. |
|
Maybe @kblondal can help review? |
|
Thanks for updating with detailed commit messages!! |
|
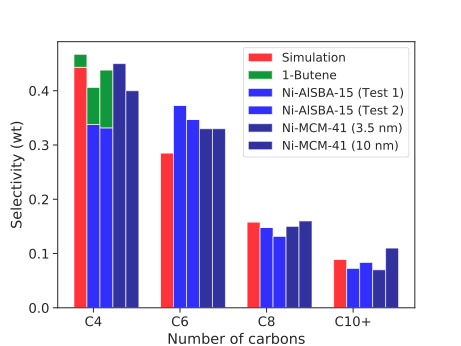
Eley-Rideal and Surface Migration families came from "Ethylene Dimerization and Oligomerization to 1-Butene and Higher Olefins with Chromium-Promoted Cobalt on Carbon Catalyst" Doi:10.1021/acscatal.7b03205. After generating a model and reproducing their experiment in cantera, I found the RMG generated model matches well with published results, with the one caveat being this was not on Cr but on a metal with -5.5 eV and -2.5 eV binding energies for carbon and hydrogen. For the other added families, I was trying to see if RMG can reproduce the mechanism found in "Mechanism of Methanol Synthesis on Cu through CO2 and CO Hydrogenation" by Grabow and Mavrikakis and added the majority of the reactions found in their mechanism. Originally, RMG had no way to react with vdW species once they adsorbed to the surface, so they'd only be able to adsorb and desorb. These new vdW families help to get rid of that dead end. Running meOH synthesis with all of the new families turned on, I am able to successfully generate a model. Thefull model can be found here |
 mjohnson541
left a comment
mjohnson541
left a comment
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I've made some general comments on this PR.
| ['FORM_BOND', '*1', 1, '*3'], | ||
| ['FORM_BOND', '*2', 1, '*4'], | ||
| ]) | ||
|
|
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Can you add:
reactantNum=2
productNum=1
here? and also for the other families?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Within the recipe or right under it? I haven't seen that in any other families yet.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
The position doesn't matter (database files are fundamentally executed like a python script so it doesn't matter). See R_Recombination.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Sorry, outside the recipe. XD
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Haha yeah, I got ya. Added those for all surface families that this commit affects.
| longDesc = u"""E0 is Ea from Xu et al. Doi:10.1021/acscatal.7b03205""" | ||
| ) | ||
|
|
||
| entry( |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
So after converting our original rate rules to training reactions only one rate rule is allowed per family. The rest need to be added as training reactions (this makes it possible to run tree generation on them).
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I think we were having issues with training reactions for surface families. I don't recall the exact problems though, it may have only been with the training reaction notebook rather than training reactions themselves.
| training set for generating rate rules to populate this kinetics family. | ||
| """ | ||
|
|
||
| # entry( |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Don't put commented information in the database. Commented stuff doesn't get loaded into the database object, so any operation that machine writes this part of the database (anytime we modify the database algorithmically) will cause all of the comments to disappear.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Right, so the story with these are that when they are uncommented, they throw errors. I think it has something to do with the fact they're the reverse of the recipe (even though other reverse training surface reactions work) but I still wanted the family to be added into the update. The error is that somehow the reaction gets turned from SurfaceArrhenius to regular Arrhenius and that's no good. The family templates do still work perfectly fine, and that only happens for training reactions.
| } | ||
|
|
||
| # Surface chemistry families that are under development and not yet working well. | ||
| surface_development = { |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I'm not sure if we already discussed this earlier, if so I apologize, but is there a reason we should be making families that are under development and are "not yet working well" available to users rather than adding them to RMG once they are working well?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I fixed this one so it is working well now.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I think Matt’s comment is about line 89 and below. A discussion worth having, but probably not as part of this PR
| Tmin = (200, 'K'), | ||
| Tmax = (3000, 'K'), | ||
| ), | ||
| rank = 1, |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Are these rates and others in this PR really known to rank 1 accuracy (experimental untuned uncorrelated within a factor of 2 accuracy)?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
No, I just made them up because they needed a rank. I trust them, the Methanol synthesis paper seems pretty legit
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
The ranks are described at https://reactionmechanismgenerator.github.io/RMG-Py/users/rmg/kinetics.html section 14.1. How they’d translate to Heterogeneous kinetics I’m not sure. But i doubt these deserve a “1”. @cfgoldsmith may have helpful experience.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
So should I just change all the ranks to 8 then? And then we can upgrade them if necessary?
| dir_list = os.listdir(fam_dir) | ||
| dir_list.remove('__pycache__') | ||
| fam_list = sorted([item for item in dir_list if os.path.isdir(os.path.join(fam_dir, item))]) # Only keep folders | ||
| print(fam_list) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Is this print necessary? If so can it be converted to a logging statement?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
This is a utility script, so there is no logging involved.
This family is is for a gas phase molecule to react directly to a surface bound molecule. Most kinetic parameters were taken from Ethylene Dimerization and Oligomerization to 1-Butene and Higher Olefins with Chromium-Promoted Cobalt on Carbon Catalyst Doi:10.1021/acscatal.7b03205 and Theoretical Investigation of the Mechanisms for Olefinic Hydrogenation on Pt(110) and Pt(111) Surfaces
This is for a surface bonded molecule to "migrate" where it is bonded to the surface. This was family found in Ethylene Dimerization and Oligomerization to 1-Butene and Higher Olefins with Chromium-Promoted Cobalt on Carbon Catalyst Doi:10.1021/acscatal.7b03205
I am moving this from development to normal surface because I have expanded the tree and added training reactions from "Mechanism of Methanol Synthesis on Cu through CO2 and CO Hydrogenation", Grabow and Mavrikakis. doi:10.1021/cs200055d
This is for a vdW bonded species that is double bonded to dissociate and each become double bonded to the surface. This is from Mechanism of Methanol Synthesis on Cu through CO2 and CO Hydrogenation", Grabow and Mavrikakis. doi:10.1021/cs200055d
This is for a single bonded surface species adding to a vdW double, triple, or quadruple bonded species and adsorbing to a surface. This was taken from "Mechanism of Methanol Synthesis on Cu through CO2 and CO Hydrogenation" by Grabow and Mavrikakis doi:10.1021/cs200055d
This is for a vdW species splitting, adsorbing to the surface, and transferring a functional group to a double, triple, or quadruple bonded surface species. This was taken from "Mechanism of Methanol Synthesis on Cu through CO2 and CO Hydrogenation" by Grabow and Mavrikakis doi:10.1021/cs200055d
This is for two vdW species reacting together and abstracting a functional group and then forming a single bond to the surface. This was taken from "Mechanism of Methanol Synthesis on Cu through CO2 and CO Hydrogenation" by Grabow and Mavrikakis doi:10.1021/cs200055d
This is for one surface species adding to the double/triple bond of another surface species causing the second to become double bonded to the surface. This is from "Mechanism of Methanol Synthesis on Cu through CO2 and CO Hydrogenation" by Grabow and Mavrikakis doi:10.1021/cs200055d
This is for the adsorption of a double/triple bonded vdW species to the surface while abstracting a functional group from a single bonded surface species causing it to become double bonded to the surface. This is from "Mechanism of Methanol Synthesis on Cu through CO2 and CO Hydrogenation" by Grabow and Mavrikakis doi:10.1021/cs200055d
Making the tree better and adding in training reactions from "Mechanism of Methanol Synthesis on Cu through CO2 and CO Hydrogenation" by Grabow and Mavrikakis doi:10.1021/cs200055d
Adding in a training reaction from "Mechanism of Methanol Synthesis on Cu through CO2 and CO Hydrogenation" by Grabow and Mavrikakis doi:10.1021/cs200055d
__pycache__ is a folder and not a family, so we don't want it to be included in the rmg_reaction_families.pdf
|
So it looks like to get this PR up to RMG-database standards for merging, RMG-Py needs to be modified so that training reactions work properly with surface families (so we don't need rate rules or uncommented training reactions). |
|
As much as I would also like this to make it into the 3.0 release, I think that Matt is correct about fixing RMG-Py to properly handle surface training reactions. Since it seems unlikely that can be done by the end of this week, I think we will need to push this to the 3.0.1 release. Would that be ok? |
|
Okay :(
… El dic. 12, 2019, a la(s) 10:38, Max Liu ***@***.***> escribió:
As much as I would also like this to make it into the 3.0 release, I think that Matt is correct about fixing RMG-Py to properly handle surface training reactions.
Since it seems unlikely that can be done by the end of this week, I think we will need to push this to the 3.0.1 release. Would that be ok?
—
You are receiving this because you were mentioned.
Reply to this email directly, view it on GitHub, or unsubscribe.
|
|
The release has been pushed back to Monday. I'm not sure if you want to try fixing the RMG-Py issues before then. |
|
lol |
This family is is for a gas phase molecule to react directly to a surface bound molecule. Most kinetic parameters were taken from Ethylene Dimerization and Oligomerization to 1-Butene and Higher Olefins with Chromium-Promoted Cobalt on Carbon Catalyst Doi:10.1021/acscatal.7b03205 and Theoretical Investigation of the Mechanisms for Olefinic Hydrogenation on Pt(110) and Pt(111) Surfaces
This is for a surface bonded molecule to "migrate" where it is bonded to the surface. This was family found in Ethylene Dimerization and Oligomerization to 1-Butene and Higher Olefins with Chromium-Promoted Cobalt on Carbon Catalyst Doi:10.1021/acscatal.7b03205
I am moving this from development to normal surface because I have expanded the tree and added training reactions from "Mechanism of Methanol Synthesis on Cu through CO2 and CO Hydrogenation", Grabow and Mavrikakis. doi:10.1021/cs200055d
This is for a vdW bonded species that is double bonded to dissociate and each become double bonded to the surface. This is from Mechanism of Methanol Synthesis on Cu through CO2 and CO Hydrogenation", Grabow and Mavrikakis. doi:10.1021/cs200055d
This is for a single bonded surface species adding to a vdW double, triple, or quadruple bonded species and adsorbing to a surface. This was taken from "Mechanism of Methanol Synthesis on Cu through CO2 and CO Hydrogenation" by Grabow and Mavrikakis doi:10.1021/cs200055d
This is for a vdW species splitting, adsorbing to the surface, and transferring a functional group to a double, triple, or quadruple bonded surface species. This was taken from "Mechanism of Methanol Synthesis on Cu through CO2 and CO Hydrogenation" by Grabow and Mavrikakis doi:10.1021/cs200055d
This is for two vdW species reacting together and abstracting a functional group and then forming a single bond to the surface. This was taken from "Mechanism of Methanol Synthesis on Cu through CO2 and CO Hydrogenation" by Grabow and Mavrikakis doi:10.1021/cs200055d
This is for one surface species adding to the double/triple bond of another surface species causing the second to become double bonded to the surface. This is from "Mechanism of Methanol Synthesis on Cu through CO2 and CO Hydrogenation" by Grabow and Mavrikakis doi:10.1021/cs200055d
This is for the adsorption of a double/triple bonded vdW species to the surface while abstracting a functional group from a single bonded surface species causing it to become double bonded to the surface. This is from "Mechanism of Methanol Synthesis on Cu through CO2 and CO Hydrogenation" by Grabow and Mavrikakis doi:10.1021/cs200055d
Making the tree better and adding in training reactions from "Mechanism of Methanol Synthesis on Cu through CO2 and CO Hydrogenation" by Grabow and Mavrikakis doi:10.1021/cs200055d
Adding in a training reaction from "Mechanism of Methanol Synthesis on Cu through CO2 and CO Hydrogenation" by Grabow and Mavrikakis doi:10.1021/cs200055d
__pycache__ is a folder and not a family, so we don't want it to be included in the rmg_reaction_families.pdf
…into new-families
|
receiving the following error for Surface reaction with HAN: |
|
I'm closing this for now because I have a newer and more updated branch to merge in after my other pull request ReactionMechanismGenerator/RMG-Py#1884 goes through |
This is a larger version of #366 with more new surface families included.
Training reactions that have been commented out throw a weird kinetics error and switch from SurfaceArrhenius to regular Arrhenius, so I've left them commented out for now. I just wanted these to make it into RMG 3.0